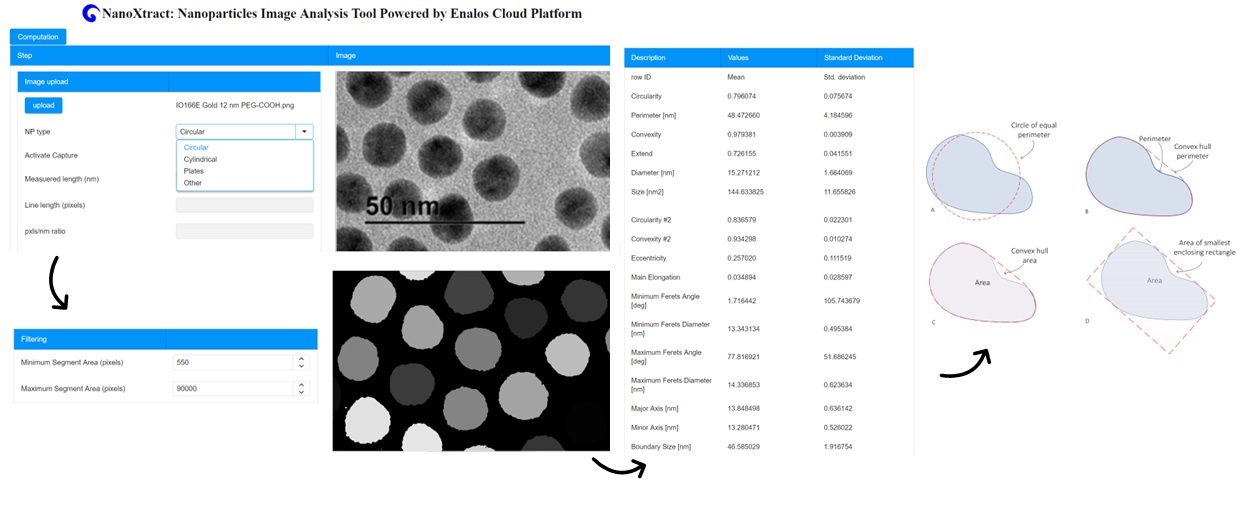
NanoXtract is a unique online tool for the calculation of 18 image descriptors based on Transmission Electron Microscopy (TEM) images of nanomaterials.

A tool for the simultaneous property and toxicity prediction of sets of chemical compounds. Users can import compounds of interest by drawing molecules and inspect a 3D visualization of their molecular structures post-drawing.
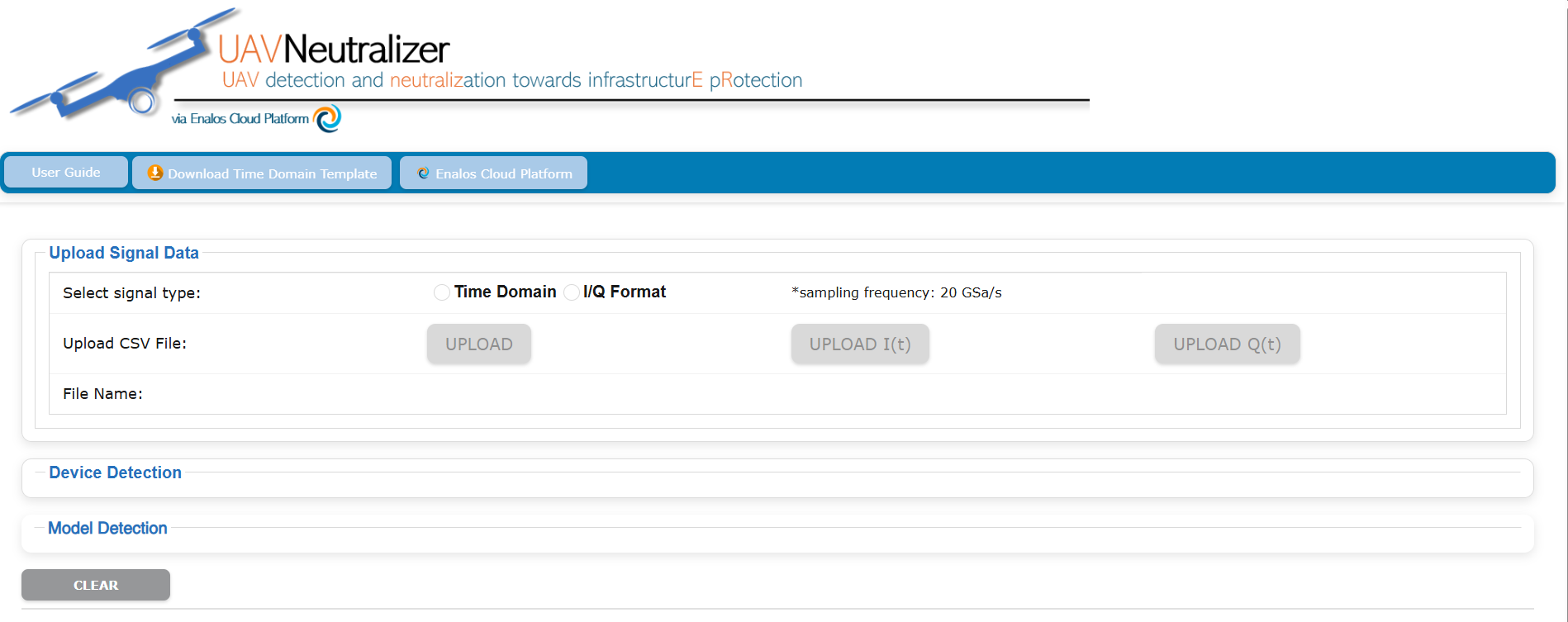
The UAVNeutralizer project aims to create an innovative, intelligent system capable of detecting and neutralizing malicious UAVs (e.g., drones), to ensure the protection of critical infrastructures. This application allows users to upload RF signals for classification into specific categories and models based on RF fingerprints.
This web application utilizes a trained machine learning model to predict the adsorption energy of NO2 molecules on nickel (Ni) clusters, for the design of novel nanocatalytic materials.
This web application utilizes a trained machine learning model to predict the adsorption energy of O2 molecules on nickel (Ni) clusters, for the design of novel nanocatalytic materials.
This web application utilizes a trained machine learning model to predict the adsorption energy of SO2 molecules on nickel (Ni) clusters, for the design of novel nanocatalytic materials.
This web application utilizes a trained machine learning model to predict the adsorption energy of CO molecules on nickel (Ni) clusters, for the design of novel nanocatalytic materials.

This web-service hosts a computational model, based on atom-attention message-passing neural networks, for the prediction of blood-brain barrier permeability.

This web-service hosts a computational model, based on atom-attention messagepassing neural networks, for the prediction of Caco-2 cell line permeability.

The MicroPlasticFate web application simulates the fate and behaviour of nano- and microplastics (NMPs) across multiple environmental scales, including regional, continental and global scale, each of which consists of compartments/media of air, soil, water and sediment.

This web service provides the functionality to predict the zeta-potential in water of individual or a set of engineered nanomaterials based on physicochemical and molecular properties.
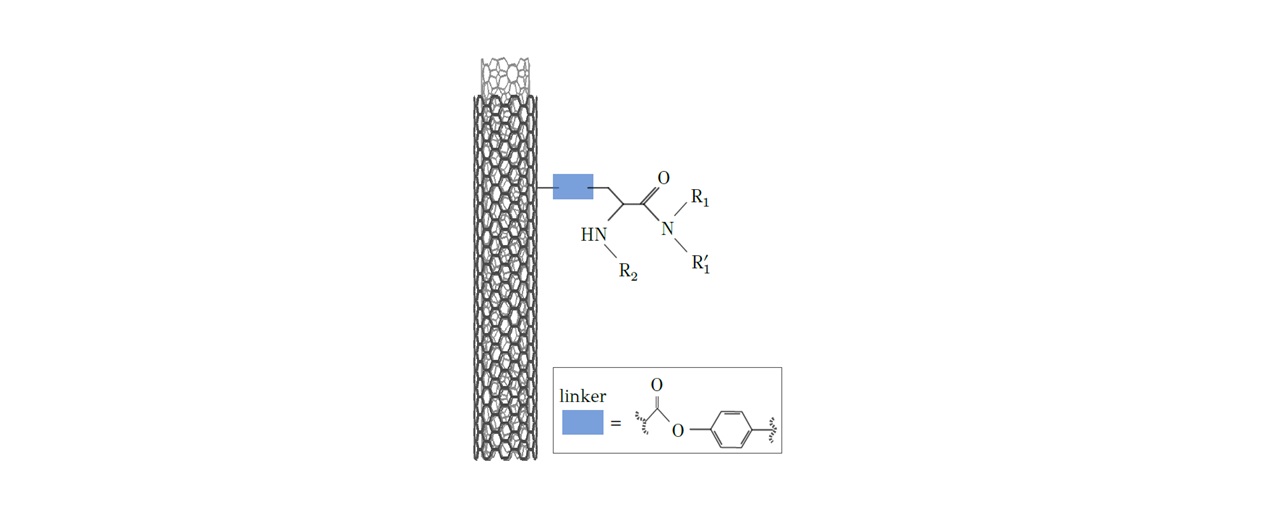
Two predictive QNAR/kNN models for the assessment of decorated multi-walled carbon nanotubes (MWCNTs) biological and toxicological profile.
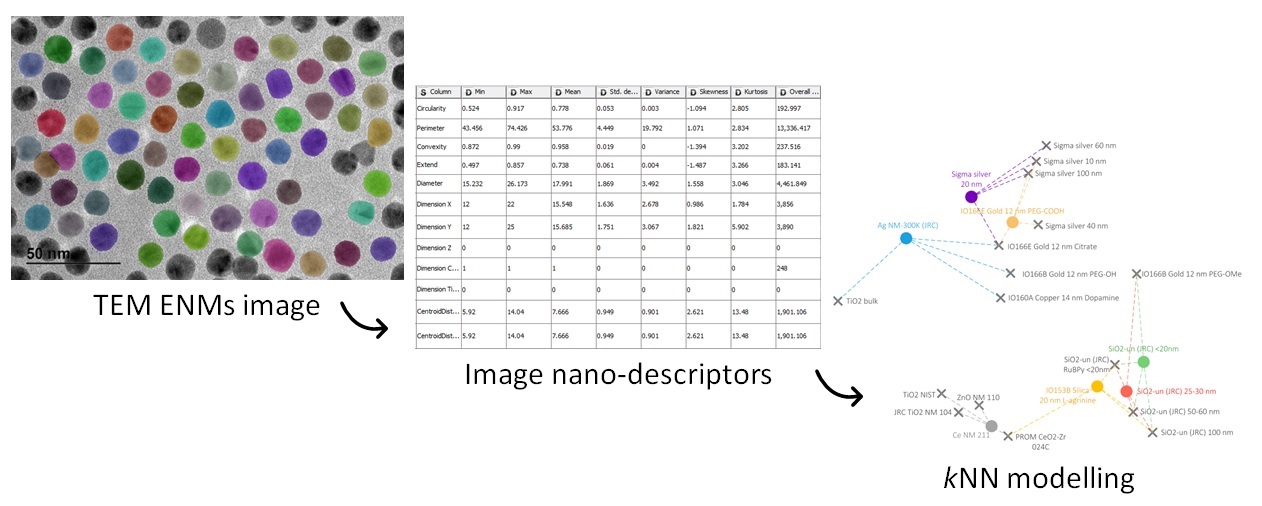
A kNN/read-across model for the prediction of nanomaterials (NMs) zeta-potential based on the NM type of core, main elongation and medium's pH value.
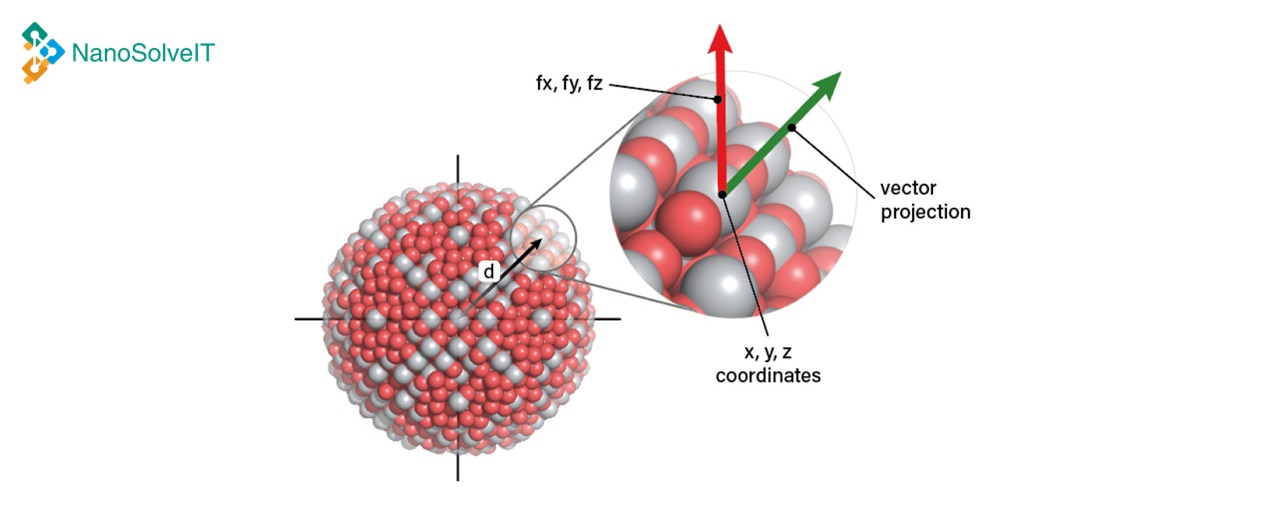
The web service provides the functionality to predict the cytotoxicity on human bronchial epithelial and murine myeloid cell lines of metal oxide nanoparticles.
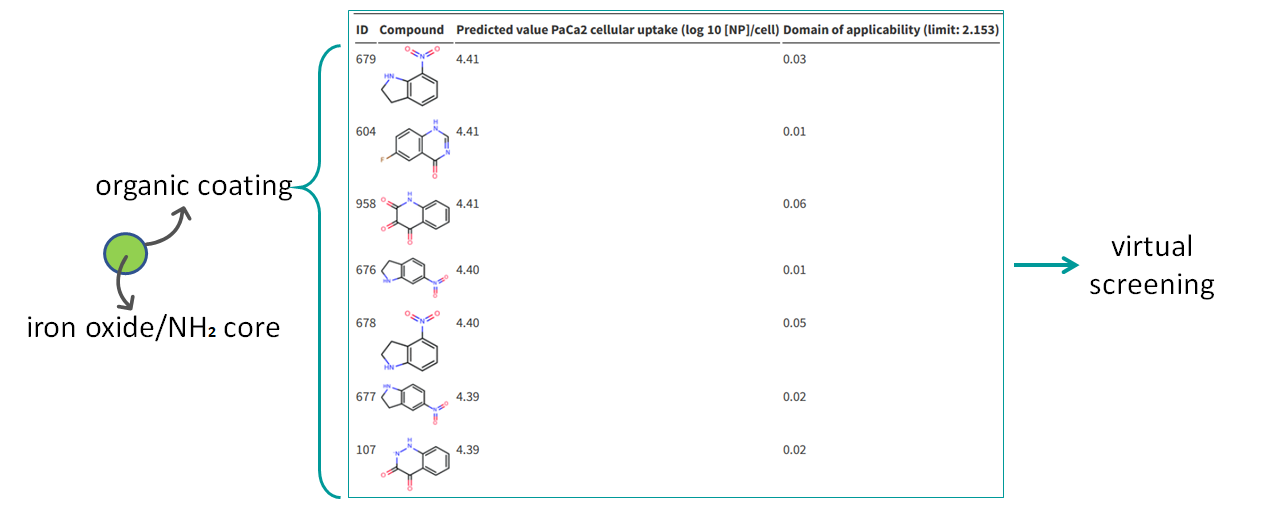
This web service hosts a QNAR model that correlates the organic surface modifiers structural descriptors of metal-oxide nanoparticles (NPs) with the same core and the NPs cellular uptakes.
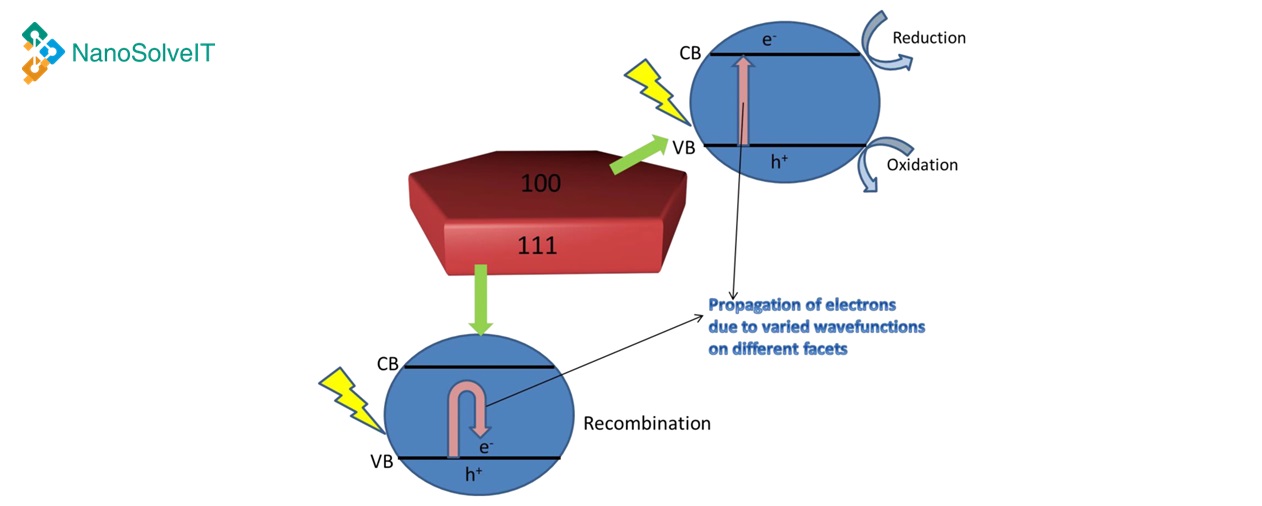
This web tool is an implementation of a model predicting the toxicity of metal oxide nanomaterials towards bronchial epithelial, murine myeloid and E. coli cell lines.

This web-service hosts a fully validated predictive model which generates toxicity predictions for iron oxide nanoparticles based on a set of five properties.
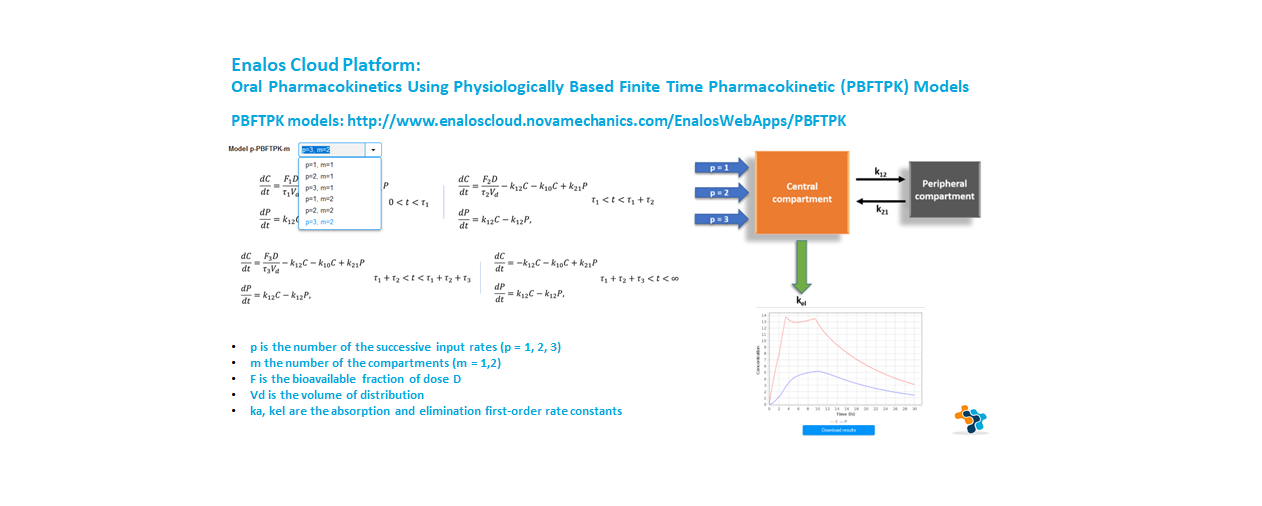
Application of physiologically based finite time pharmacokinetic (PBFTPK) models for the analysis of oral drug absorption phenomena, based on the concept of finite absorption time.

The MouseTox web-service hosts a model that predicts the cytotoxic effects of small molecules to NIH/3T3 (mouse embryonic fibroblast) cells, based on calculated structural descriptors.
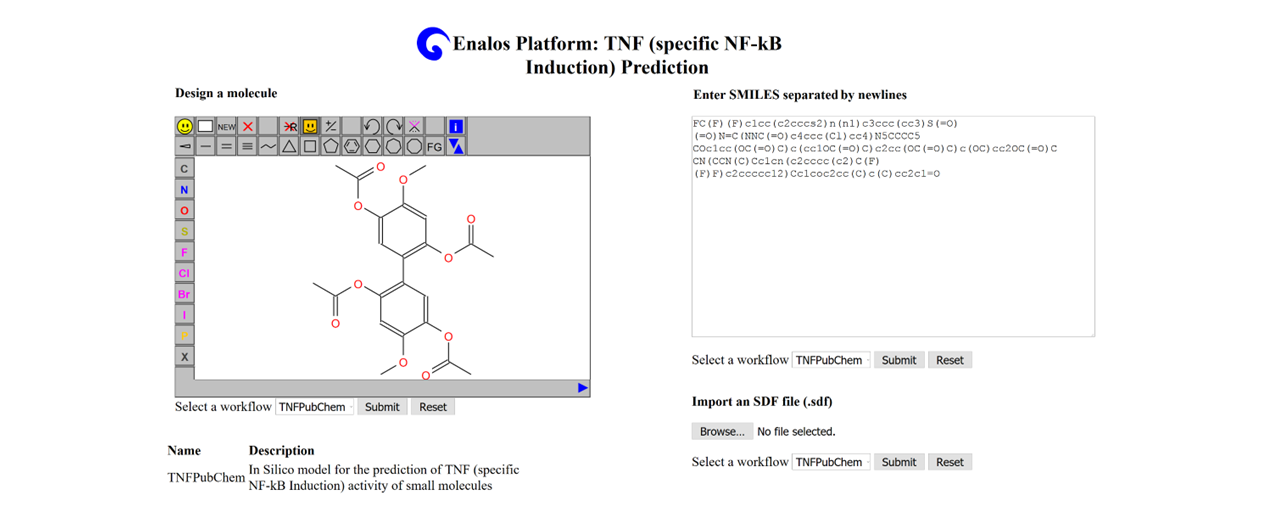
This web-service hosts the Tumor Necrosis Factor TNFPubChem in silico model for the prediction of TNF (specific NF-kB Induction) activity of small molecules.
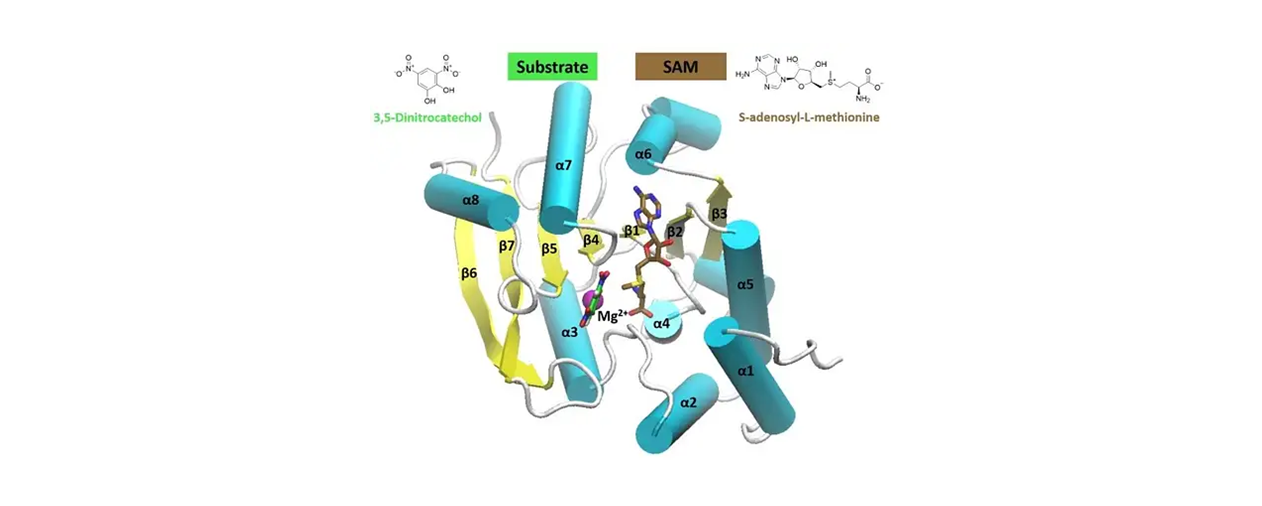
This web service hosts a predictive consensus model to facilitate the design and virtual screening of novel potent small-molecule inhibitors of COMT (catechol-o-methyl transferase).
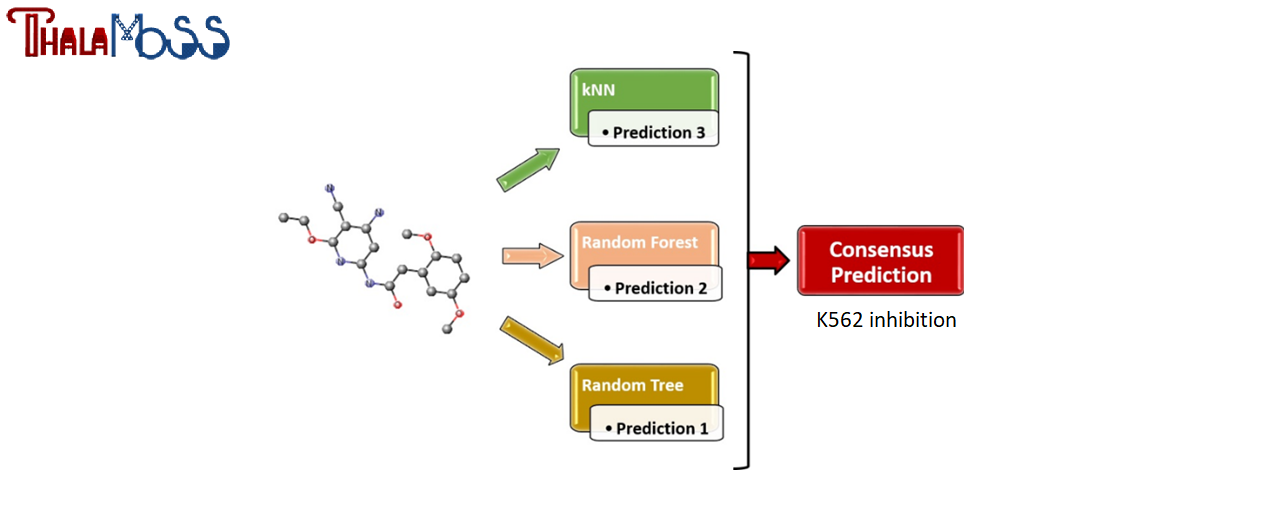
The web-service hosts a consensus predictive model for the assessment of K562 inhibition. This model contributes to the identification of compounds that possibly have therapeutic action against β-thalassemia.
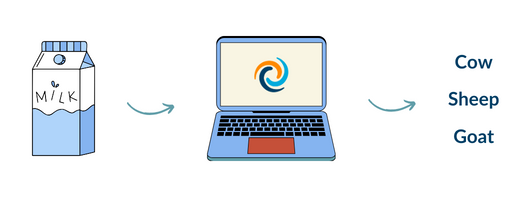
This web service hosts a validated model to predict the milk type (cow, goat, sheep) of various samples based on a series of milk properties.

This web service hosts a validated Support Vector Machine regression model to predict the mass loss of metal compounds to measure sample’s corrosion resistance under the salt spray test.
This web-service hosts a predictive model from the drug discovery perspective, targeting new HbF inducers to explore new therapies for thalassemia patients.
This web service hosts a validated model to predict the chelation outcome for thalassemia patients by applying clustering-based and machine-learning-based stratification techniques to a comprehensive clinical dataset from thalassemia patients.
This web-service hosts a consensus in silico predictive model for the assessment of the ability of small molecules to inhibit the cytotoxic effect of the Tumor Necrosis Factor (TNF).
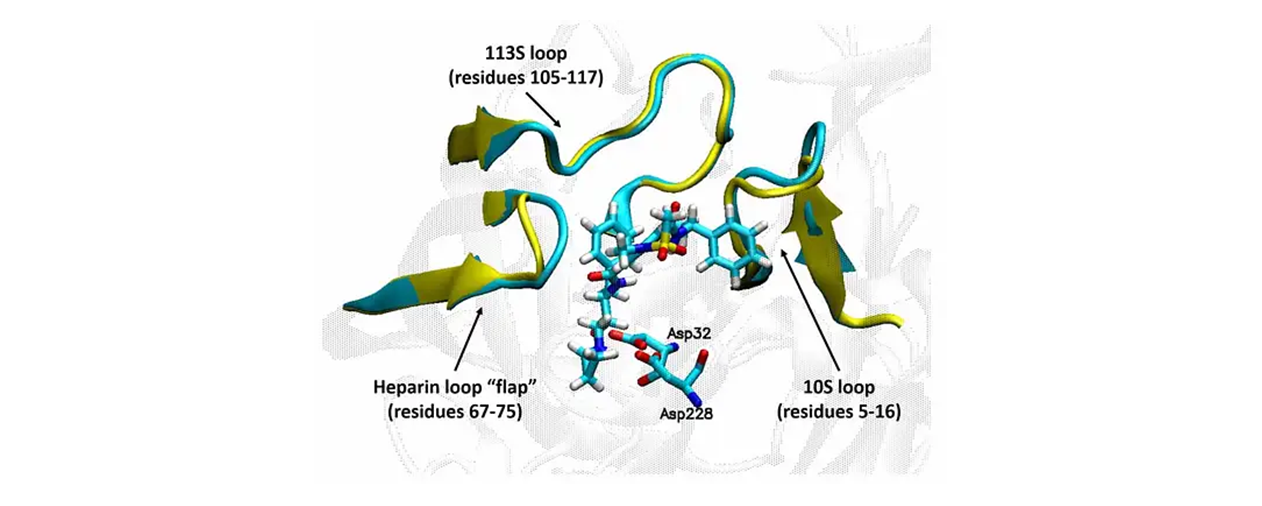
This web service hosts a robust, validated according to OECD principles ligand-based predictive model for beta-site APP-cleaving enzyme (BACE) inhibitory data.
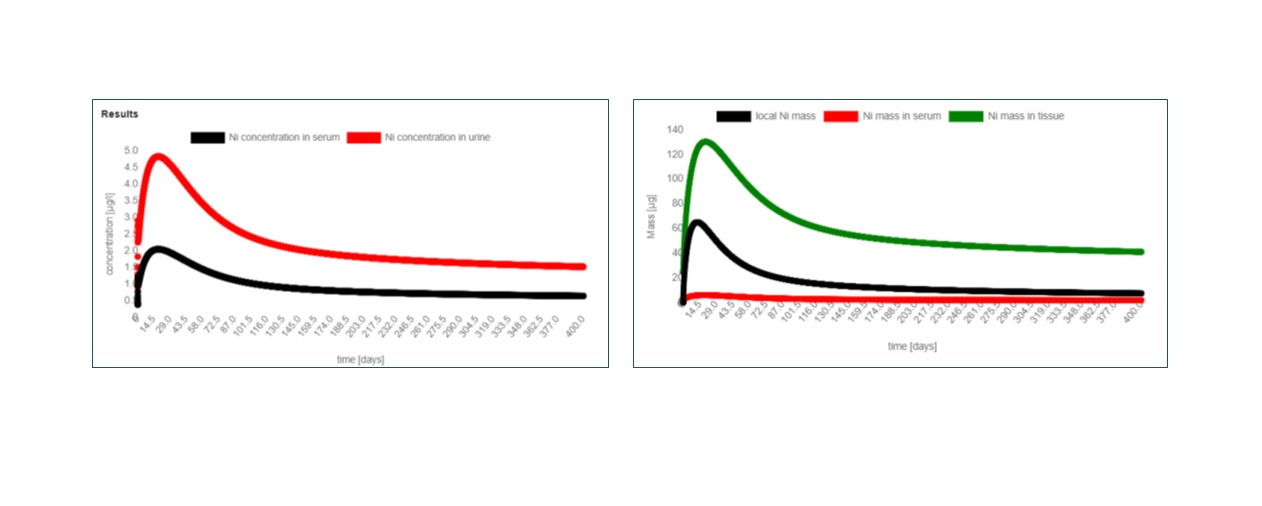
This tool is used for the prediction of the rate at which Nickel (Ni) is released from cardiovascular devices following implantation of the body, using the biokinetics model proposed by Saylor et al.